aws4SPM - Structure adaptive smoothing in SPM
Introduction
A very popular software tools for analyzing functional Magnetic Resonance Imaging data is SPM by the Functional Imaging Laboratory at the Wellcome Department of Imaging Neuroscience. In order to enhance the signal-to-noise ratio it provides the possibility to smooth the data in a pre-processing step by a Gaussian filter. However, this comes at the cost of reducing the effective resolution, which is especially disturbing at high spatial resolutions. In a series of recent papers [1, 2, 4, 6] we have shown, that using a structural adaptive smoothing algorithm based on the Propagation-Separation method allows for signal detection while preserving the shape and spatial extent of the activation areas. Here, we provide our implementation of this algorithm [1] as a toolbox for SPM.
The application of a Gaussian filter to the three dimensional images of the fMRI scan as implemented in SPM has mainly two effects. Firstly, if activation has spatial extent adjacent voxel will contain similar time courses and thus the averaging lowers the noise level while conserving the signal. Secondly, it increases the smoothness of the random t-field and thus lowers the number of independent tests as well as the corresponding global thresholds. However, with Gaussian smoothing we can achieve a reduction in the signal-to-noise ratio at the cost of a possible bias at the border of the activated regions only. This leads to an increased power of the tests and therefore more sensitivity and specificity of signal detection in all voxel except in a neighborhood of the border of the region. The size of this neighborhood depends on the amount of smoothing that is applied. In this neighborhood specificity of signal detection may decrease. The optimal amount of smoothing depends on both signal strength and spatial extent.
In [1] we designed an algorithm for structure adaptive smoothing fMRI data based on the Propagation Separation approach. The idea behind structural adaptive smoothing is to use information on the structures of interest in the smoothing algorithm for locally modeling the data. If, for example an anatomical image is disturbed by noise, the interesting structures may be the different tissues, which are characterized by a certain grey level value. Thus locally the data can be described by a constant. This information can be used in our structural adaptive smoothing algorithm, which preserves shape and border of this interesting structures. For fMRI the structures of interest are not found in a single three dimensional image of the time series, but in the value of the BOLD parameter or the estimated effect. We expect this parameter to differ significantly from zero in activated areas and to be consistent with zero elsewhere. [1] therefore suggested to apply the structural adaptive smoothing to the Statistical Parametric Map after estimating the parameters from the linear model. This smoothing algorithm is implemented in this toolbox
Our Project
The toolbox has been originally developed by Devy Hoffmann and Karsten Tabelow at the Weierstrass Institute for Applied Analysis and Stochastics within the Project "F10 - Image and signal processing in the biomedical sciences: diffusion weighted imaging - modeling and beyond" of the DFG Research Center MATHEON. However, the original version was not well integrated into the standard workflow of SPM and installation required replacing some original SPM Matlab files (sic!). Chiara D'Alonzo and Karsten Tabelow therefore re-wrote the toolbox to fit into the SPM toolbox API.
Note
This toolbox comes with absolutely NO WARRANTY! It is not intended for any purpose! It is especially not intended for any clinical use, but for evaluation purpose only.
Related software
Here, we want to point the attention towards an implementation of the structure adaptive smoothing algorithm as described in [1] in R Language and Environment for Statistical Computing which has more features and is under constant development. The package is called fmri and freely available for download at CRAN as well as at NITRC.org.
We also refer to the "Adaptive Smoothing"-plugin which is available in BrainVoyager QX version 2.8.2.
Download
Here, you can download the current (beta-)version after the re-work. Note, that the use of older version (up to 0.4.3) is highly depreciated! This also means, that documention in [5] is no longer appropriate!
| aws4SPM toolbox (re-work version 1.0.1): | Download (30kB) |
| aws4SPM toolbox (version 1.0.2 - including Win64 binary now!): | Download (40kB) |
Getting started
The toolbox aws4SPM requires Matlab and SPM2 to be installed on your computer as well as a MEX-C-Compiler, if you are not using on of the OS where we provide pre-compiled binaries (Linux, Mac, Windows 64bit). After downloading the toolbox archive, extract the files into your SPM toolbox directory. The archive contains a C-file "smAd.c" which has to be compiled before using (if your system does not match one of those, we provide binaries for). In order to do this launch Matlab. Start the compilation by entering the command
mex smAd.c
Now the installation is complete and you can start SPM and use the toolbox.
Documentation
In order to use the toolbox, you will need the SPM.mat file generated by the Estimate job (See [7], page 71).
You can also directly use the SPM.mat file produced by the Specify 1. level step (See [7], page 61),
without having to explicity start the Estimate job.
In this case you wil be prompted to accept to estimate the model and AWS4SPM will take care of that too.
Refusing of estimating the model will result in terminating the AWS job without any result.
Since version 1.0.1 it is not necessary anymore to change directory to the one containing the SPM.mat file
before starting AWS4SPM.
It is important not to use the smoothed image volumes while specifying the model in the Specify 1. level step. Just skip the Smooth step or do not use the produced files, usually starting with the dafault prefix "s". (See [7], page 57)
To begin, open MATLAB and launch the SPM toolbox. Open the batch editor by clicking on the Batch button. (screenshot)
The batch editor window will open. Select MPM → Tools → AWS4SPM. (screenshot)
In the editor, select the SPM.mat file and change adaptation (lambda) and maximal (hmax) bandwidth, if you wish to.
Run the batch clicking on the now green small triangle (play).
If the model has been estimated (or after that has been done in AWS), you will be prompted to select a contrast.
If it is the first time you run AWS with this SPM.mat file, you will not see any contrast
and will have to define a new one. Only contrasts defined in AWS4SPM are visible in the contrast manager started by the toolbox,
even if you have already defined a contrast calling the Results job.
On the other hand contrasts defined and used in AWS4SPM are only visible running the AWS batch
and not in the contrast manager called by the Results button.
You can define as many new contrasts at a time as you want, but you can choose only one to be processed. You can see and use the others, if you restart the batch.
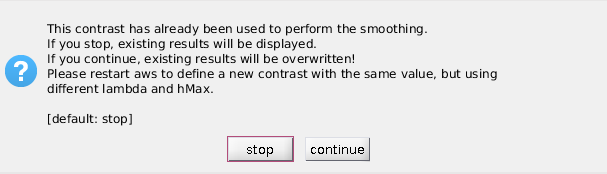
Each contrast processed in AWS is then saved with the corrispondent lamba and hmax values.
If you want to reuse a contrast with another bandwidth specification, you will have to define it again (restarting the batch)
or accept to overwrite previous results. Selecting an existing contrast can be also used to rerun the Results part on an already processed contrast.
If you select an existing contrast, you will be prompted to express your choice.
(screenshot)
Now the AWS4SPM will start and the smoothing adaptative algorithm implemented in smAd.c will be executed. On the MATLAB console you can see how the performed steps proceed.
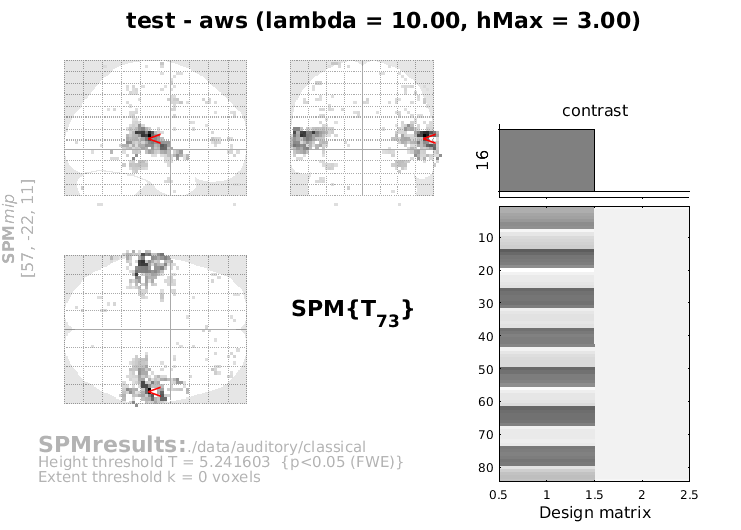
Once the calculations are done, the Results part is internally started (as usual you will have to specify mask, p-value, threshold, etc.) and the results will be visualized.
You can ask us, if you need assistance: Mail to Karsten Tabelow.
Changelog
13.10.2016 Version 1.0 released: major changes from 0.x.x version to better integrate the toolbox in the SPM workflow
19.10.2016 Version 1.0.1 released: bugfix - directory problem solved when using a model not yet estimated
02.01.2017 Version 1.0.2 released: Happy New Year version including a binary for Win64 (thx to Siawoosh Mohammadi!).
Literature
- K. Tabelow, J. Polzehl, H.U. Voss, and V. Spokoiny. Analyzing fMRI experiments with structural adaptive smoothing procedures, NeuroImage 33(1), 55-62 (2006).
- J. Polzehl, K. Tabelow. fmri: A package for analyzing fmri data, RNews 7(2), 13-17 (2007).
- H.U. Voss, K. Tabelow, J. Polzehl, O. Tchernichovski, K. Maul, D. Salgado-Commissariat, D. Ballon, and S.A. Helekar, Functional MRI of the zebra finch brain during song stimulation suggests a lateralized response topography, Proceedings of the National Academy of Sciences (PNAS) 104(25), 10667-10672 (2007).
- K. Tabelow, J. Polzehl, A. M. Ulug, J. P. Dyke, R. Watts, L. A. Heier, and H. U. Voss. Accurate Localization of Brain Activity in Presurgical fMRI by Structure Adaptive Smoohting, IEEE Trans. Med. Imaging , 27(4), 531-537 (2007).
- D. Hoffmann, K. Tabelow. Structural adaptive smoothing for single-subject analysis in SPM: the aws4SPM-toolbox, WIAS Technical Report No. 11,(2008)
- K. Tabelow, V. Piech, J. Polzehl, and H. U. Voss. High-resolution fMRI: Overcoming the signal-to-noise problem, Journal of Neuroscience Methods 178(2), 357-365 (2009).
- The FIL Methods Group (and honorary members), SPM12 Manual.
Karsten Tabelow Last modified: Mon Jan 02 13:06:55 CEST 2017